a Public Health Unit, South Western Sydney Local Health District.
b Faculty of Health, University of Technology, Sydney, New South Wales, Australia.
c Division of Microbiology, SydPath, St. Vincent’s Hospital, Sydney, New South Wales, Australia.
d The iThree Institute and School of Medical and Molecular Biosciences, University of Technology, Sydney, New South Wales, Australia.
e Microbiology Department, South Eastern Area Laboratory Service (SEALS), Prince of Wales Hospital, Randwick, New South Wales, Australia.
f School of Medical Sciences, The University of New South Wales, Sydney, New South Wales, Australia.
g Department of Microbiology, Children’s Hospital at Westmead, New South Wales, Australia.
h Department of Microbiology and Infectious Diseases, Royal Prince Alfred Hospital, Camperdown, New South Wales, Australia.
Correspondence to John Ellis (email:John.Ellis@uts.edu.au).
To cite this article:
Fletcher S et al. Descriptive epidemiology of infectious gastrointestinal illnesses in Sydney, Australia, 2007–2010. Western Pacific Surveillance and Response Journal, 2015, 6(4):7–16. doi:10.5365/wpsar.2015.6.2.006
Objective: There is a lack of information about the prevalence of gastrointestinal illnesses in Australia. Current disease surveillance systems capture only a few pathogens. The aim of this study is to describe the epidemiology of infectious gastrointestinal illnesses in Sydney, Australia.
Methods: A retrospective cross-sectional study of patients with gastrointestinal symptoms who visited tertiary public hospitals in Sydney was conducted between 2007 and 2010. Patients with diarrhoea or loose stools with an enteric pathogen detected were identified. Demographic, clinical and potential risk factor data were collected from their medical records. Measures of association, descriptive and inferential statistics were analysed.
Results: In total, 1722 patients were included in this study. Campylobacter (22.0%) and Clostridium difficile (19.2%) were the most frequently detected pathogens. Stratified analysis showed that rotavirus (22.4%), norovirus (20.7%) and adenovirus (18.1%) mainly affected children under 5 years; older children (5–12 years) were frequently infected with Campylobacter spp. (29.8%) and non-typhoid Salmonella spp. (24.4%); infections with C. difficile increased with age. Campylobacter and non-typhoid Salmonella spp. showed increased incidence in summer months (December to February), while rotavirus infections peaked in the cooler months (June to November).
Discussion: This study revealed that gastrointestinal illness remains a major public health issue in Sydney. Improvement of current disease surveillance and prevention and control measures are required. This study emphasizes the importance of laboratory diagnosis of enteric infections and the need for better clinical data collection to improve management of disease risk factors in the community.
Gastrointestinal (GI) illnesses are a significant public health problem, resulting in one third of working Australians missing on average one day of work each year.1 GI illnesses are a burden to the health-care system, costing approximately 1.2 billion Australian dollars annually.2,3 In Australia, the national disease surveillance system captures only campylobacteriosis, typhoid fever, giardiasis and salmonellosis; however, campylobacteriosis is not reportable in New South Wales (NSW), the largest state. In NSW, medical practitioners and hospitals are required to report notifiable conditions to the local public health units (PHU) on the basis of reasonable clinical suspicion. Pathology laboratories are required to notify a positive result for specified infectious diseases and medical conditions. Primary schools and childcare centres are encouraged to seek advice from their local PHU when they suspect an infectious disease outbreak in their centres using standard reporting forms from the Australian Department of Health.4–7
The PHU in the state of NSW are responsible for investigating reports of enteric disease based on established reporting requirements, they then enter the data into the state-wide NSW Notifiable Conditions Information Management System. Outbreaks are detected through a variety of sources including notifiable diseases surveillance data, reports from general practitioners, institutions or laboratories and the public.4–6
The surveillance data reveal that enteric viruses, mainly norovirus and rotavirus, are the most common causes of non-food GI illness, accounting for approximately 15–18% of all GI illness cases in NSW.4,5 One study showed that approximately 25% of all cases of gastroenteritis are foodborne with an estimated 4.1 million foodborne gastroenteritis cases occurring in 2010. Pathogenic Escherichia coli, norovirus, Campylobacter and non-typhoid (N-T) Salmonella were responsible for over 93% of foodborne illness from known pathogens. However, the majority of cases (80%) did not have a known pathogen identified.8,9
Previous studies revealed that approximately 30% of people will seek medical attention for GI illness;10,11 among this group, only about 20% (range: 14–27%) will have confirmatory tests with stool specimens.12 In addition, only a few selected pathogens are reportable to the infectious disease surveillance system. Therefore, several emerging and re-emerging pathogens cannot be captured.13 Previous reports indicated that a significant proportion of illnesses were not reported in the surveillance system and that the majority of pathogens causing illness remain unknown.8,9 This creates a paucity of information about the prevalence of GI illnesses in Australia. This study described the clinical and epidemiological characteristics and the common pathogens associated with GI illnesses in Sydney, Australia in 2007–2010.
A retrospective cross-sectional study was conducted on patients who presented to the four public referral hospitals or affiliated clinics in Sydney with GI symptoms and had an enteric organism detected in their stool from January 2007 to December 2010. Hospitals in this study were selected by convenience sampling. Cases were randomly selected using an online random number generator (StatTrek, Atlanta).
Demographic details (age, gender, post code, country of birth, relationship status and language spoken); clinical data (vital signs, date of onset, date of hospitalization, date of hospital separation, symptoms, diagnosis, organism[s] detected, treatment received and co-morbidities [surgery, HIV/AIDS, cancer, transplant]); and potential risk factor data (antibiotic use/chemotherapy, chronic GI illness, consumption of suspect food, men who have sex with men [MSM] status and travel history) were collected from the patients’ medical records.
Laboratory results for all stool specimens that tested positive for an enteric organism were collected from 2007 to 2010 except for one hospital that only included data from 2008 to 2010. Patients with diarrhoea (liquid or watery stools taking the shape of the container) or loose (unformed) stools were identified from the laboratory records provided by the hospitals.
The laboratory methods for the diagnosis of enteric organisms have been previously described.14,15 Tests for fungi or other pathogens were conducted only by special requests from clinicians.
Briefly, all laboratories conducted testing for adenovirus and rotavirus routinely in all children aged 5 or younger unless otherwise indicated or requested by the clinician. Rotavirus, adenovirus serotypes 40 and 41 and norovirus were detected by either an enzyme immunoassay (EIA), or the RIDA® Quick Rotavirus/Adenovirus Combi immunochromatographic test and the RIDASCREEN® norovirus test (R-Biopharm Inc., Darmstadt, Germany). All tests were conducted following the manufacturer’s recommendations.
Bacteria identification was routinely performed in all laboratories using standard culture methods. In summary, selective media were used: Xylose Lysine Deoxycholate agar was inoculated for the detection of Salmonella, Shigella and Yersinia; Aeromonas, Plesiomonas and Vibrio spp. on Horse Blood Agar; Campylobacter spp., Campylobacter agar and Clostridium difficile on C. difficile agar, Oxoid Australia. C. difficile was detected using EIA for hospitalizations greater than three days or if otherwise indicated (e.g. history of antibiotic use, chemotherapy or immuno-suppressed patients).
All hospitals processed stools by a wet preparation in saline and examined for white blood cells, red blood cells and cysts, ova and parasites (COP). Direct microscopy was routinely performed on all stool specimens for the detection of COP and concentration techniques were performed on request at some hospitals. Techniques included a modified iron haematoxylin stain incorporating carbol fuchsin to enhance the detection of acid-fast Isospora, Cryptosporidium, Cyclospora, and direct DNA extraction using a QIAamP DNA stool minikit (Qiagen, Hilden, Germany) for the identification of Entamoeba spp, as previously described.16 EIA was performed as a screening test for Giardia intestinalis, Cryptosporidium parvum and Entamoeba histolytica. All positive findings from the EIA were confirmed by microscopy.
Based on previous literature,17 we estimated that each laboratory receives approximately 10 000 specimens per year over the study period and the prevalence of uncommon microbes is approximately 5% for diarrhoeal cases. A sample size of 436 was required for each study site at a 95% confidence level with 80% power and 2% margin of error. Oversampling of cases was performed to avoid any shortfalls in missing medical records.
Descriptive analysis was done for demographic characteristics. The association between demographic characteristics, clinical symptoms, pathogens detected and potential risk factors was examined using the Pearson’s chi-squared test. Associations between potential risk factors (age group, surgery, transplant, HIV/AIDS, cancer, chronic GI illness, antibiotic use, travel history, consumption of suspect food and MSM status) and selected pathogens (Blastocystis spp, Dientamoeba fragilis, Campylobacter, C. difficile, N-T Salmonella and Shigella) were placed into a binary logistic regression model. A backward stepwise multiple logistic regression was conducted for those selected pathogens with variables having a P-value lower than 0.25 in the univariate analysis.18 Subjects with missing variables were excluded from the regression model. Odds ratios (OR) and 95% confidence intervals (95% CI) for the association were reported. Statistical analyses were performed using SPSS version 18.0 (International Business Machines Corp, New York, USA).
This study received ethical approval from the Human Research Ethics Committees for each of the four hospitals and the University of Technology, Sydney and was guided by the Australian National Statement on Ethical Conduct of Research involving humans.
Four public referral hospitals were included in the analysis.
Of the 19 490 patients with diarrhoea or loose stools at the four selected hospitals, 1722 cases were included in this study (Figure 1). The recruitment of cases at Hospital D was lower than expected due to administrative issues. For Hospital C, only the medical records between January 2008 and December 2009 period were reviewed and the laboratory results between January 2008 and December 2010 were included, whereas the other hospitals covered the period between January 2007 and December 2010.
* Multiple stool specimen counted.
Participants were aged between 25 days and 99 years (mean: 28.3 years, standard deviation [SD]: 29.5 years). The majority of the participants at Hospitals A and C were in the age groups older than 12 years (67%), while children under 5 years were predominantly seen at Hospitals B (72%) and D (42.2%) (Table 1). The overall mean length of stay in hospital was 8.9 days (SD: 21.4 days) and this increased with age. Patients aged 50–75 years (mean: 20.3 days, SD: 30.4 days) and those 75 years and older (mean: 18.2 days, SD: 18.5 days) had a longer length of stay compared with children under 5 years (mean: 4.3 days, SD: 16.7 days) and 5–12 years (mean: 4.3 days, SD: 10.3 days).
Campylobacter spp, (22.0%), C. difficile (19.2%) and N-T Salmonella (14.0%) were the most frequently detected bacteria. The most frequently detected viruses were norovirus (10.7%), rotavirus (9.4%) and adenovirus (7.6%); Blastocystis spp. (7.2%), G. intestinalis (2.9%) and D. fragilis (1.8%) were the most common enteric protozoa detected. Other organisms were detected in less than 3% of cases.
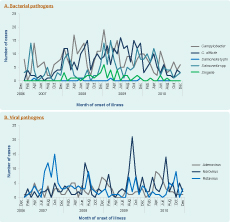
Except for Campylobacter and N-T Salmonella spp., which increased in summer months (December to February), no seasonal patterns were found for infections, with bacterial pathogens (Figure 2, Panel A). In contrast, viral infections, which predominantly affected children under 5 years, showed clearer seasonal patterns (Figure 2, Panel B). Rotavirus and norovirus activities peaked in the cooler months (June to October and July to September, respectively); adenovirus showed a less consistent trend.
Overall, Campylobacter infection was associated with watery diarrhoea (49.1%), abdominal pain (63.1%) and fever (56.5%; P < 0.01). N-T Salmonella infection was significantly associated with vomiting (60.8%), abdominal pain (59.2%) and fever (66.3%; P < 0.01). The majority of people infected with norovirus, rotavirus and adenovirus infections reported vomiting and lethargy (P < 0.01) (Table 2).
Children under 5 years (n = 637) presented with diarrhoea (97.8%; P > 0.05), vomiting (66.7%; P = 0.001), lethargy (56.5%; P < 0.001) and fever (body temperature > 37.8 °C; 56.0%; P < 0.05). Older children aged 5–12 years (n = 168) had similar clinical symptoms except that they reported higher rates of abdominal pain (72.0%) and fever (64.9%); however, these symptoms were not statistically significant (P > 0.05) when compared with other age groups. The most frequently detected GI pathogens in the less than 5 years age group were rotavirus (22.4%), norovirus (20.7%), adenovirus (18.1%), Campylobacter spp. (15.1%) and N-T Salmonella spp. (12.3%). Children aged 5–12 years were also frequently infected with Campylobacter spp. (29.8%) and N-T Salmonella spp. (24.4%) (Table 2).
FBI, foodborne illness; MSM, men who have sex with men; and N-T, non-typhoid.
* Information available for cases from Hospital C only (n = 313).
Patients older than 12 years mainly presented with diarrhoea (range: 99–100%) and abdominal pain (range 27–76%). Common pathogens infecting people in the 13–24 years and 25–49 years age groups were Campylobacter spp. (42.7% and 30.9%, respectively) and N-T Salmonella spp. (21.3% and 16.3%, respectively); those 50 years or older were predominantly infected with C. difficile (50–75 years, 46.3% and > 75 years, 53.9%). Sixty-nine per cent of people infected with C. difficile were 50 years or older (P < 0.001). Infections with Blastocystis spp. also increased with increasing age (P < 0.01). Overall, infections with enteric viruses decreased with increasing age (P < 0.01), although G. intestinalis was highest in children aged 12 years or younger (P < 0.01) (Table 2).
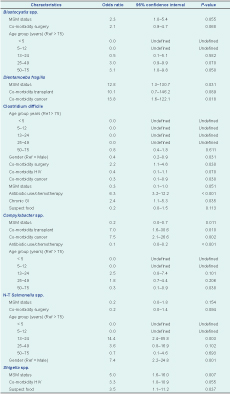
In multivariate analyses, MSM status was associated with Shigella spp., (OR = 5.0; 95% CI: 1.6–16.0), Blastocystis (OR = 2.3; 95% CI: 1.0–5.4) and D. fragilis (OR = 12.8; 95% CI: 1.3–130.7). Infection with C. difficile was associated with prolonged antibiotic therapy (OR = 6.3; 95% CI: 3.2–12.2), recent surgery (OR = 2.2; 95% CI: 1.1–4.6) and chronic GI illness (OR = 2.4; 95% CI: 1.1–5.3) (Table 3).
GI, gastrointestinal illness; MSM, men who have sex with men; and Ref, reference group.
To our knowledge, this is the largest multihospital study to describe the epidemiology of infectious GI illnesses in NSW, Australia. We provided an overview of GI illnesses associated with GI pathogens among people seeking care in Sydney across four major public hospitals.
There are 53 public hospitals in the eight local health districts in the Sydney Metropolitan Area, and four (8%) were included in this study to represent high density population centres. Clinical laboratories within the selected hospitals provide laboratory services for smaller hospitals in their local health districts and for some rural health services in the Newcastle, Illawarra and Hunter regions. This captures a wide population of NSW.
Viral gastroenteritis had a distinct seasonal pattern with rotavirus and norovirus infections peaking in the cooler months; adenovirus showed a less consistent monthly trend. These seasonal trends have been previously described in Sydney14 and other settings19,20 and is useful for public health planning and resource allocation. Whereas infections with Campylobacter and N-T Salmonella spp. were mainly foodborne, both appeared to have occurred more frequently in warmer months in the study. However, the seasonal difference was not statistically significant, probably due to small sample size. Increased incidence of viral gastroenteritis in cooler months and bacterial illnesses in warmer months implies that health promotional messages should be developed to target the respective high risk groups in each season. The relatively high prevalence of antibiotic-associated C. difficile infections suggests that existing protocols and practices for the control of C. difficile should be carefully reviewed and modified where necessary.
For parasites, Blastocystis was the most common parasite detected in symptomatic patients in this study; in contrast, a previous study found Giardia and Cryptosporidium to be the main intestinal parasites associated with enteric infections in Australia.21 This study only detected Giardia and Cryptosporidium in only 3% and 1% of cases, respectively. Previous literature revealed that Blastocystis spp. have emerged as the most commonly detected enteric protozoa in developed settings.22 Despite much controversy about the pathogenicity of Blastocystis spp., several reports have described their association with abdominal pain, persistent diarrhoea and irritable bowel syndrome-like symptoms,23–25 and other reports postulate that pathogenicity may be subtype dependent.26 D. fragilis, an emerging protozoan pathogen, was found in 3% of cases. The combination of conventional and molecular diagnostics has led to the increased detection of D. fragilis in Australia with its prevalence rivalling Giardia in developed settings.24,27,28
This study found that GI illnesses affected people of all ages; however, the clinical symptoms and the prevalence of GI pathogens varied across different age groups. There were slightly more males than females in this study, which is in contrast to Australian national data which suggest an overall higher rate of GI illness in females, especially in the 20–40 years age group.9 The reason for these differences is not clear, but it may be related to differences in exposure between males and females at different stages of the lifespan. For example, a study from the United States of America found that more males than females will seek medical attention for severe GI symptoms.12
Children were more likely to be infected with enteric viruses, especially rotavirus, norovirus and adenovirus, as previously described in NSW.2,14,15 However, older patients were more likely to be infected with C. difficile as also described in Australia29 and elsewhere.30,31 In this study, older patients (aged 50 years or above) had longer lengths of stay in hospital compared with younger children. Dysfunction of the immune system with aging and co-morbidities may increase the length of stay.32,33 The increased risk of C. difficile infection associated with prolonged antibiotic use and particularly among people with extended length of stay indicates a need for good antibiotic stewardship. Existing protocols should be carefully reviewed and modified where necessary.34
There was a significant association between infection with Shigella spp., HIV/AIDS and MSM, which warrants further investigation. Shigella spp. are easily transmitted via faecal-oral sexual contact,35 and outbreaks linked to unsafe sexual practices have been described among MSM,36 a high-risk group for HIV/AIDS in Australia.37 Public health education and promotion could be targeted toward high risk groups.
This study, like most retrospective studies, has some limitations. Only symptomatic cases that had a positive laboratory test were included in this study which may bias the results because for asymptomatic cases, the likelihood of patients reporting to hospitals is low. Obtaining clinical information from asymptomatic cases is difficult. Also, reporting to hospital for a microbiological test would be strongly influenced by the location of the hospitals and whether or not testing facilities are conveniently located in relation to their routine activities. Current clinical guidelines for the management of acute gastroenteritis do not recommend routine collection and testing of stools; hence, the results cannot represent the full spectrum of community acquired gastroenteritis.
The hospital data were reviewed retrospectively. Incompatible data records among hospitals prevented analysis of some risk factors. Also, information on some potential risk factors (e.g. MSM status, HIV/AIDS diagnosis and diarrhoea) may have been incomplete and may have affected the results.
Only some enteric pathogens are included in testing protocols. As a result, some known pathogens such as Staphylococcus aureus and Bacillus cereus, which are likely to cause foodborne outbreaks,6 were not tested in most stool specimens. Sensitivity of some of the tests such as microscopy and EIA15,28,38 are limited and some cases may be missed.14,36,38 Also, stool testing protocols differ among hospitals. The immuno-chromatographic test used by one hospital detected all adenovirus serotypes, not just the enteric serotypes 40 and 41; hence, a positive result does not necessarily mean the serotype found was the cause of the GI illness. In addition, testing for norovirus at some hospitals mainly occurred when outbreaks were suspected, which may have resulted in selection bias.
This study has revealed that GI illness is a major public health issue in Sydney, Australia with implications for resource management and disease surveillance and control. The study has identified various risk factors that can be addressed by public health interventions. Information on disease risk factors is essential for the control of infectious diarrhoea and should be routinely collected in a systematic way across hospitals. The consistent use of well-organized electronic medical records is recommended.
None declared.
None.
The authors would like to acknowledge all the laboratory and medical records staff at the four hospitals who facilitated data collection.