a National Institute of Hygiene and Epidemiology, Ha Noi, Viet Nam.
Correspondence to Le Quynh Mai (email: lom9@hotmail.com or lom9@nihe.org.vn).
To cite this article:
Le TT et al. Circulation of influenza B lineages in northern Viet Nam, 2007–2014. Western Pacific Surveillance and Response Journal, 2015, 6(4):17–23. doi:10.5365/wpsar.2015.6.1.022
Introduction: Influenza B viruses circulate throughout Viet Nam, and their activities vary by region. There have been two antigenically distinct lineages of influenza B viruses co-circulating in the past 20 years; however, only one lineage is selected as a component of contemporary trivalent seasonal influenza vaccines. To improve the understanding of circulating influenza B lineages and influenza vaccine mismatches, we report the virus lineages circulating in northern Viet Nam over an eight-year period (2007–2014).
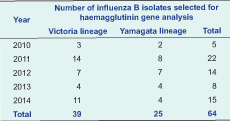
Methods: Lineages of 331 influenza B viruses were characterized by haemagglutination inhibition assay against standard reference ferret (Yamagata) and sheep (Victoria) antisera. Sequence analysis of the haemagglutinin gene was performed in 64 selected influenza B isolates.
Results: The proportion of influenza B lineages changed by year. The Yamagata lineage predominated in 2007, 2008 and 2012; the Victoria lineage predominated in 2009–2014 except 2012. The two lineages showed continuous evolution over time. The Northern Hemisphere’s influenza vaccine components were mismatched with the predominant circulating viruses in 2007, 2009 and 2014.
Discussion: The seasonality of influenza B activity is more variable in tropical and subtropical regions than in temperate zones. Our data showed a common co-circulation of both influenza B lineages in northern Viet Nam, and it was difficult to predict which one was the predominant lineage. Quadrivalent influenza vaccines containing both lineages may improve the effectiveness of influenza vaccine programmes in the future.
Influenza infection occurs as an annual seasonal epidemic in winter or early spring in countries with temperate climates.1 Currently, four antigenically distinct groups of influenza viruses have been identified as the cause of human infection, including two subtypes of influenza A (A/H1N1 and A/H3N2) and two lineages of influenza B. The two influenza B lineages are represented by the reference strains B/Victoria/2/87 and B/Yamagata/16/88. They have co-circulated with influenza A viruses since 1983.2 The proportion of the two B lineages varies by year and country; however, current seasonal influenza vaccine only includes one influenza B strain. As the two lineages have no cross-reactivity, the decision for vaccine lineage selection can be difficult in years when both influenza B lineages are circulating.3 Furthermore, differences in evolutionary and epidemiological dynamics between the Victoria and Yamagata lineages can confound the selection.4
In Viet Nam, influenza constitutes an important cause of influenza-like illness (ILI) among outpatients seeking clinical care.5 Influenza viruses circulate year-round with two distinct peaks in virus circulation6 unlike in temperate climates where a single peak in the winter season is typical. Moreover, the climates of southern and northern Viet Nam differ remarkably. The climate in northern Viet Nam is humid and subtropical, while southern Viet Nam has a tropical climate all year round. Transmission patterns of influenza vary considerably in the two regions.7 The patterns of influenza B virus in Viet Nam did not appear synchronous with seasonal influenza A viruses. Influenza A viruses peak in the spring usually in February and March. Influenza B viruses peak from November to March in the north, are detected at similar levels throughout the year in the southern region and are at much higher levels in November to May in the central region.6
The Viet Nam National Influenza Surveillance System (NISS) was established in 2005 based on sentinel sites in four regions (northern, southern, highlands and central). The National Influenza Center (NIC) at the National Institute of Hygiene and Epidemiology, Ha Noi (NIHE) conducts influenza virological surveillance in northern Viet Nam. The surveillance data provides information on the effect and seasonality of influenza in Viet Nam and monitors influenza virus strains circulating throughout the country.5,6
The two influenza B lineages have co-circulated and caused seasonal outbreaks in Viet Nam and the Asia-Pacific region since 1987;8 however, laboratory-based surveillance and detailed analyses of viral transmission patterns have not been conducted previously. In this study, we report the circulating lineages of influenza B in Viet Nam in the years 2007 to 2014 to improve the knowledge about this circulating virus.
Subjects of all ages presenting to one of the seven sentinel sites in northern Viet Nam (two central hospitals in Ha Noi, three district hospitals and two outpatient clinics)5 with ILI using the World Health Organization (WHO) definition (body temperature ≥ 38 °C plus cough and/or sore throat) within three days of onset were included in the study.8
Nasopharyngeal swabs (NPS) or throat swabs (TS) were collected by trained nurses using cotton swabs (Hanacomedical Co., Ltd., Saitama, Japan). Samples were collected from the first two ILI patients per day on weekdays. Swabs were stored in in-house viral transport media.3,8 Samples were transferred on Friday or Monday of the following week on ice to the NIC for virological testing.6
The NPS and TS influenza B positive samples by reverse transcription polymerase chain reaction were selected for viral isolation according to NISS protocols. Viruses were harvested and stored at –80 °C. Influenza B isolates were subtyped using the haemagglutination inhibition assay (HI) with reference antigens and antiserum of B/Victoria and B/Yamagata lineages using the WHO reagent kit. Ferret or sheep sera (pre-treated with receptor-destroying enzymes [Denka Seikan Co., Ltd., Tokyo, Japan]) were raised against reference strains representing the B/Victoria lineage (B/Brisbane/32/2002) and B/Yamagata (B/Shanghai/361/2002). HI assays were performed in 96-well micro-titre plates with 0.5% chicken erythrocytes cells. Reference antiserum was diluted from 1:10 to 1:1280. Influenza B viruses were diluted to 4 haemagglutinin (HA) units/25µl and tested following WHO guidelines.9 The lineages of influenza B isolates were identified by comparing them with both reference antisera; the higher titre is assumed to be homologous to Victoria or Yamagata lineage.
The characterized influenza B strains were compared with the strains of WHO-recommended vaccine components for the Northern and/or Southern Hemispheres to check if the influenza B lineage was matched each year from 2007 to 2014. Mismatches between influenza B strains and vaccine strains of the same lineages were noted when their HI titre differences were more than twofold.
The influenza B isolates with sufficient 8 HA units were selected for HA genetic analysis by sequencing at NIC-NIHE. RNA extraction was conducted on 140μl aliquot of each isolate using the viral RNA extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The RNA was transcribed to cDNA using the influenza A virus universal primer (Uni 12) AGC AAA AGC AGG as described.10 The HA gene was amplified with segment-specific primers for influenza B with primers HA-25F:ATC CAC AAA ATG AAG GCA and HA1140R: ACC AGA ATA GCT CCGA. The PCR products were purified with PCR purification kit (Qiagen, Valencia, CA, USA) and labelled with Big Dye Terminator v3.1 cycle sequencing kits (Applied Biosystems, Waltham, MA, USA) according to manufacturer’s instruction and then analysed by an ABI 3100 automatic DNA sequencer.
Sequences were assembled using Lasergene analysis software, version 8.0 (DNASTAR, Inc., Madison, WI, USA). Multiple sequences alignment was conducted with CLUSTAL-X (Conway Institute University College Dublin, Dublin 4, Ireland) for the major coding regions of HA segments.11 Phylogenetic trees of the HA sequences were constructed by the maximum likelihood (ML) method with bootstrapping (1000 replicates), referencing the HA genes of strains B /Brisbane/32/2002 (B/Victoria), B/Jiangsu/10/2003 (B/Yamagata) and strains from the National Center for Biotechnology Information (NCBI) influenza virus resource website.12 ML trees were estimated using the best fit nucleotide substitution model.13 To quantify amino acid sequence diversity, Basic Local Alignment Search Tool (BLAST) in Molecular Evolutionary Genetics Analysis (MEGA) 5 was used to search within NCBI to find the closest sequence available for representative strains of each lineage.11
All sequences reported in this study have been deposited in the GenBank database under accession numbers from KT359277 to KT359340.14
The National Institute of Hygiene and Epidemiology, Viet Nam and Centers for Disease Control and Prevention, Atlanta, Georgia, United States of America provided ethical committee approval for the study. All participants provided written informed consent.
In total, 331 influenza B isolates were collected from NISS and we selected 64 virus isolates that had HI titres higher than 8 HA units for sequence analysis (Table 1).

Influenza circulating patterns Data from NISS in the years 2007 to 2012 indicated that influenza B circulated throughout the year with activities primarily peaking in March and April (Figure 1).

PCR, polymerase chain reaction.
For the 331 influenza B isolates, we found 195 isolates belong to the Victoria lineage and 136 isolates belong to the Yamagata lineage. They were detected in all years of the study. The Yamagata lineage was predominant in 2007, 2008 and 2012. The Victoria lineage was predominant in 2009 through 2011 and 2013 through 2014 (Table 2).
Note: B/Malaysia/2506/2004-like virus and B/Brisbane/60/2008-like virus belong to the B/Victoria /7/87 lineage. B/Florida/04/2006-like virus, B/Wisconsin/1/2010-like virus and B/Massachusetts/2/2012-like virus belong to the B/Yamagata/16/88 lineage.
The HI titres of the reference antisera against most of the influenza B/Victoria lineage study isolates (190/195, 97.3%) were less than twofold different from the WHO-recommended vaccine strain (B/Brisbane/60/2008). Similarity, almost all influenza B/Yamagata isolates (124/136, 91.2%) reacted well with the reference antisera raised against the recommended vaccine strains (B/Florida/04/2006; B/Wisconsin/01/2010 or B/Massachusetts/2/2012).
The HA gene phylogenetic analysis of the 64 virus isolates revealed that 39 isolates belonged to the Victoria lineage and 25 isolates to the Yamagata lineage. The phylogenetic trees showed different genetic diversities of Victoria and Yamagata lineages based on nucleotide differences in the HA1 region. The influenza B/Victoria phylogenetic tree can be diversified into two clades (V1 and V2). Clade V1 contained all of the isolates in 2010 (3 isolates), 2011 (14 isolates) and almost all isolates in 2012 (6/7 isolates). Clade V1 was clustered with isolates from Australia, Bangladesh and Thailand. They all shared amino acid changes at S134P, N165K and A199T compared to the recommended vaccine strain (B/Malaysia/2506/2004). In total, 16 isolates collected in 2012 (1 isolate), 2013 (4 isolates) and 2014 (11 isolates) were grouped into Clade V2. They were highly homologous with viruses from Australia, Thailand, the United States of America and the reference B/Brisbane/32/2002 strain (Figure 2).
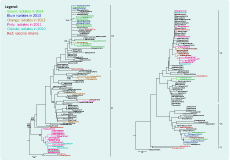
The Yamagata lineage was split into three clades as Y1, Y2 and Y3. Clade Y1 did not contain any of the 25 influenza B/Yamagata isolates in our study. Clade Y2 was grouped by 17 isolates in 2010–2012, two isolates in 2014 and others from China, the United States of America and Japan. Clade Y2 was closely related to the recommended vaccine B/Florida/4/2006 strain. The common amino acids different from Clade Y2 to reference strain B/Jiangsu/10/2003 are at R48K, P108A, I150S, I166N, T182A, S203N and D230G. Clade Y3 had four isolates in 2013 and two isolates in 2014 together with circulating viruses in China, Thailand and the United States of America. In 2009–2012; this clade showed amino acid differences at N116K, K299E and E313K when compared to the B/Jiangsu/10/2003 stain (Figure 2).
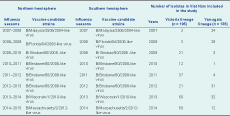
For most of the study years of 2007 through 2014 (6/8, 75.0%), the same influenza B vaccine candidate strains were recommended for both the Northern and Southern Hemispheres, except 2009 and 2012. Vaccine formulation in the Northern Hemisphere was updated in 2009 from B/Malaysia/2506/2004 to B/Brisbane/60/2008 (Victoria lineage). The Yamagata lineage was also updated in 2012 from B/Florida/04/2006-like to B/Wisconsin/1/2010 and B/Massachusetts/02/2012 in 2014.
Possible mismatches were found between the Vietnamese predominant influenza B circulating strains and the WHO-recommended influenza B vaccine components (Northern Hemisphere) in 2007 (B/Yamagata lineage), 2009 and 2014 (B/Victoria lineage) (Table 2).
Furthermore, the WHO-recommended strains only matched about half of the contemporary circulating influenza B strains in 2008–2012. For example in 2012 (B/Yamagata lineage), only 31/52 (59.9%) isolates in this study were grouped as Yamagata lineage (Table 2).
Co-circulation of both influenza B lineages with different proportions by year is common not only in subtropical countries such as Viet Nam and Hong Kong Special Administrative Region (SAR), China, but also in temperate countries such as the United States of America and European countries.1,3,15,16 This is the first one about circulation of influenza B lineages in Viet Nam.
Our data indicated that the pattern of circulating influenza B lineages varies, similar to the findings in Australia, Brazil, Hong Kong SAR, China and Scotland.17–20 Mismatches were found between the vaccines and circulating strains throughout the study years, suggesting the vaccines may not have been effective for northern Viet Nam in those years.
Our study found no differences in titres in most of the circulating viruses against the reference, indicating that these viruses were not antigenically distinguishable from reference and vaccine candidate strains. There was no evidence of major antigenic drift of the influenza B viruses during the study period. Therefore, the influenza B isolates in this study still shared most of their antigenic properties with the vaccine-candidate strains.
Phylogenetic analysis showed both Victoria and Yamagata lineages have high similarity to viruses circulating in Viet Nam and neighbouring countries. The genetic diversity of the HA gene indicated at least two subclades within each lineage, and the presence of amino acid substitutions in HA epitopes of isolates indicated antigenic drift is ongoing (Figure 2).
Influenza B viruses, unlike influenza A viruses, have multiple evolutionary lineages which can co-exist for considerable periods of time.21 This has occurred since the early 1980s when a new lineage (B/Yamanashi/16/88-like) appeared to evolve from B/USSR/100/83-like viruses. Since then it has co-circulated with the existing virus lineage (B/Victoria/2/87-like).2 According to data from the WHO Global Influenza Surveillance and Response System, both lineages of influenza B viruses have co-circulated in different countries concurrently,3,22 making the selection of annual influenza vaccine components for influenza B difficult.
Although influenza B viruses are less likely to trigger widespread epidemics than influenza A, influenza B viruses co-circulate with influenza A and sometimes are predominant in Viet Nam. The prevention of influenza infection remains a public health priority in Viet Nam; vaccination is the main tool for prevention. Trivalent vaccine has been used worldwide, but it may not be effective if the influenza B vaccine components are mismatched. Recently, the quadrivalent influenza vaccine that contains two influenza B strains was used in some countries such as Canada, United States of America and several European countries.23 The efficacy of quadrivalent vaccines was found to be higher than that of the trivalent ones,24 although its effectiveness is yet to be determined in Viet Nam. Quadrivalent influenza vaccines may be one solution to help improve the efficacy of influenza vaccine programmes in the future. Meanwhile, changes of influenza B strains in the upcoming influenza seasons remain unpredictable. It is recommended that influenza surveillance be continued year-round for monitoring the disease trends to help inform influenza vaccine policies for disease prevention and control.
This study has several limitations. As the sample size is small, the overall trends of influenza B circulation may be difficult to discern without further analysis. Circulating patterns of the two influenza B lineages may not be representative of all of Viet Nam as the study only included samples from northern Viet Nam. Future studies that include variables from different geographical regions as well as climate, social conditions and other factors are encouraged. Information for other genes such as neuraminidase (NA) and internal gene segments is lacking. Nevertheless, results from another study has shown that the evolutionary and epidemiology dynamics observed in NA and internal gene segments were similar to those observed in the HA genes in both the Victoria and Yamagata lineages.4
Our results provide additional information about virological characteristics of seasonal influenza B viruses in northern Viet Nam, which may lead to new influenza vaccine policies in the future.
None declared.
None.
We would like to thank Dr Juliet Bryant for scientific editing of this manuscript. We thank the National Institute of Hygiene and Epidemiology for their continuous support for our work. We thank NISS, supported by WHO and United States Centers for Disease Control and Prevention for providing the study data.