a Papua New Guinea Institute of Medical Research, Goroka, Papua New Guinea.
b Center of Excellence for Influenza Research and Surveillance, St Jude Children’s Research Hospital, Memphis, United States of America.
c Papua New Guinea National Agriculture Quarantine and Inspection Authority, Port Moresby, Papua New Guinea.
Correspondence to Paul Horwood (paul.horwood@pngimr.org.pg).
To cite this article:
Jonduo M et al. Surveillance of avian influenza viruses in Papua New Guinean poultry, June 2011 to April 2012. Western Pacific Surveillance and Response Journal, 2013, 4(4):11–15. doi:10.5365/wpsar.2013.4.4.004
We investigated the circulation of avian influenza viruses in poultry populations throughout Papua New Guinea to assess the risk to the poultry industry and human health. Oropharyngeal swabs, cloacal swabs and serum were collected from 537 poultry from 14 provinces of Papua New Guinea over an 11–month period (June 2011 through April 2012). Virological and serological investigations were undertaken to determine the prevalence of avian influenza viruses. Neither influenza A viruses nor antibodies were detected in any of the samples. This study demonstrated that avian influenza viruses were not circulating at detectable levels in poultry populations in Papua New Guinea during the sampling period. However, avian influenza remains a significant risk to Papua New Guinea due to the close proximity of countries having previously reported highly pathogenic avian influenza viruses and the low biosecurity precautions associated with the rearing of most poultry populations in the country.
Influenza virus is a major respiratory pathogen that infects an average of 5−15% of the global population each year, with approximately 500 000 human deaths related to influenza annually.1 Currently all known influenza A viruses are naturally maintained in aquatic birds.2 Occasionally these influenza viruses of avian lineage cross natural species barriers and infect other susceptible bird species and/or mammals including humans, pigs and horses. The interspecies transmission of highly pathogenic avian influenza (HPAI) virus to poultry populations often results in devastating disease outbreaks.
In 1996, a HPAI strain of H5N1 emerged in South-East Asia and extended throughout several Asian, Middle Eastern, African and European countries. Its re-emergence in 2003 resulted in the death of more than 62 million birds in Thailand alone, almost half of which were backyard poultry.3 Death caused by infection and preventive measures (such as depopulation) implemented to control the spread of the HPAI H5N1 virus resulted in considerable socioeconomic burdens for many of the affected countries.4 The recent emergence of a novel H7N9 virus in China (March 2013) has increased fears about the spread of influenza viruses with pandemic potential from poultry populations.5 The transmission of these viruses over long distances by migrating birds is a concern for countries such as Papua New Guinea that have large poultry populations with few biosecurity precautions.
Poultry production accounts for 45% of the total annual livestock production in Papua New Guinea, and poultry consumption is second only to pigs.6 The short turn-around time, ease in rearing, market demand and high income from poultry production makes it more profitable than most other livestock rearing in Papua New Guinea. Most poultry farming in the country is conducted in semi-enclosed areas or free-ranged village settings. Relatively few poultry farms are commercialized and therefore do not have high biosecurity settings to reduce potential introduction of influenza viruses into the poultry population. The free-ranged village/backyard chickens are often raised together with other animals within the same pen (e.g. pigs and ducks). The village chickens also have unrestricted access to water and feed sources that may be utilized by wild birds, thus increasing the risk of exotic disease transmission.
In this paper we report a cross-sectional study to determine the presence of circulating avian influenza viruses and the seroprevalence of neutralizing antibodies to avian influenza viruses in poultry populations across Papua New Guinea.
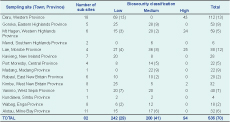
Oropharyngeal swabs, cloacal swabs and serum were obtained from 536 poultry (466 chickens and 70 ducks) from 82 sub-sites within 14 selected provinces from June 2011 to April 2012 (Table 1 and Figure 1). Qualified field officers from the Papua New Guinea National Agriculture Quarantine and Inspection Authority carried out the sampling during their routine surveillance programme, adhering to the guidelines of the Food and Agriculture Organization of the United Nations (FAO) for avian sampling.7

* Samples in brackets were from ducks (unknown species) with the remaining from chickens.
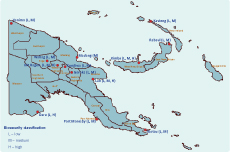
Sampling was conducted in three types of biosecurity settings: high, medium and low. These classifications were based on the amount of exposure the sampled poultry population had to other birds and/or animals. Thus, poultry sites with little-to-no exposure to other animals or birds were classified as high (e.g. commercial farms); sites with some exposure were classified as medium (e.g. semi-enclosed farms); and sites with unlimited exposure were classified as low biosecurity containment (e.g. free-range village chickens).
Oropharyngeal swabs, cloacal swabs and serum were obtained from poultry and sent at 4 °C to the laboratory for analysis. Upon arrival at the laboratory, the samples were stored at –80 °C (–20 °C for sera) until required for analysis. Total RNA was extracted from oropharyngeal and cloacal swabs using the QIAamp Viral RNA Minikit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The extracted RNA was tested for the presence of influenza A virus by real time reverse-transcriptase polymerase chain reaction (PCR) assays supplied by the Centers for Disease Control and Prevention (Atlanta, GA, USA). Samples positive or equivocal for avian influenza viruses were further tested for influenza A/H5 and A/H7 using previously published assays.8 Aliquots of all samples were sent to the Center of Excellence for Influenza Research and Surveillance, St Jude Children’s Research Hospital (Memphis, TN, USA) for isolation and subtyping of avian influenza virus isolates.
A total of 36 paired oropharyngeal and cloacal samples collected from farms and provinces that had samples deemed equivocal were passaged three times in 10-day old embryonated chicken eggs. A sample was considered negative for isolation if no virus was isolated upon three passages. For increased sensitivity in detection of viral genome, deep-sequencing was also performed on the equivocal samples. Briefly, viral RNA was extracted, transcribed to cDNA and subjected to whole-genome amplification according to previously published methods.9 The resulting PCR products were then library-prepped and sequenced on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using the paired-end sequencing chemistry. After removal of MiSeq indices, analysis was performed using CLC Genomics Workbench 6.5 (CLC bio, Aarhus, Denmark) using the following process: for quality trimming sequence reads were filtered at the quality-limit threshold of 0.05; short reads and reads with more than two ambiguous bases were removed. Remaining reads were then de novo assembled using the fast-contig mapping mode at the minimum contig length of 200 base pairs; paired-reads were aligned using the scaffold option. Assembled contigs were then subjected to BLASTn search against the National Center for Biotechnology Information (Bethesda, MD, USA) database for viral sequences.
Sera were analysed for the presence of influenza A virus antibodies using the IDEXX AI MulitS Screen ELISA (IDEXX Laboratories, Rydalmere, Australia), according to the manufacturer’s instructions. All serum samples were individually tested on three separate occasions to ensure the validity of results.
Influenza A virus was not detected in any of the oropharyngeal or cloacal swabs (n = 536 each). Four samples had results recorded as equivocal as crossing-threshold values of 36–40 were detected. These samples were tested for influenza A/H5 and A/H7 using real-time PCR; however, all of the samples were negative. Further analysis of these samples using egg inoculation and next-generation sequencing at St Jude Children’s Research Hospital (Memphis, TN, USA) resulted in no detection of influenza A virus.
Despite all serum samples being tested on three independent occasions, influenza A antibodies were not detected in any of the samples. Positive and negative control reactions supplied with the kits confirmed the validity of the results.
This paper is the first to investigate the presence and distribution of avian influenza viruses in poultry populations in Papua New Guinea. Influenza virus and antibodies were not detected in any of the samples, suggesting that there is low (or no) circulation of avian influenza viruses in poultry in the country. Poultry and wild bird surveillance programmes in other countries, such as Australia and New Zealand, have also found low prevalence of circulating avian influenza viruses.10
The failure to detect avian influenza viruses in poultry does not necessarily mean that Papua New Guinea is at low risk for an outbreak. The recent detection of H5N1 in West Papua (Indonesia)11 is a concern for Papua New Guinea as this region shares a land border with West Papua.The recent outbreak of Newcastle Disease virus in poultry populations in the north-west region of Papua New Guinea12 highlights the potential for the incursion of exotic diseases into this region. Indeed the maiden outbreak of chikungunya was first detected in this region13 before subsequently spreading throughout much of the country.
Papua New Guinea is in close proximity to South-East Asian countries endemic for the H5N1 and H7N9 viruses.3,5 The spread of these viruses through the migration of waterfowl may be a potential source of incursion into non-endemic areas.14 Although wild bird surveillance studies have shown that there is a low prevalence of avian influenza viruses in Australia, and an absence of HPAI,10 avian influenza introduction from this direction is also a possibility given the nomadic migration of some duck species between Australia and Papua New Guinea.15 H5N1 has not been reported in the Pacific region since its re-emergence in 2003, despite being detected in the West Papua province of Indonesia. Previous studies have suggested that the Pacific region is protected from the incursion of HPAI influenza viruses by the uncommon migration of waterfowl across Wallace’s Line.16 However, it is important that active surveillance continues so that outbreak mitigation steps can be rapidly implemented in the event of incursion of these viruses. In particular, future surveillance studies should focus on wild waterfowl and the potential for the introduction of avian influenza viruses through migration and nomadic movements of these birds.
In this study we report that there is no evidence of avian influenza circulation in Papua New Guinean poultry populations. However, there are some limitations to this study. A cross-sectional analysis for avian influenza viruses may not be sufficiently sensitive when a low prevalence of virus is circulating. The short lifespan of poultry bred for meat and the low number of samples collected from each site may have contributed to the non-detection of avian influenza viruses and antibodies. Therefore, it is recommended that long-term sentinel surveillance should be established at sites where there is a risk of avian influenza introduction, such as sites close to border crossings and lakes used by waterfowl.
Although wild waterfowl migration routes are unlikely to be the source of exotic avian influenza introduction, the landborder with West Papua (Indonesia) and the poultry husbandry practices in Papua New Guinea mean that there is still a relatively high risk of introduction into the country. The introduction of HPAI viruses into Papua New Guinea could create a huge socioeconomic burden. Poultry provides the only source of protein consumption for many people in rural regions, and a large outbreak may have far-reaching health implications. Poor diagnostic capacity at a national level17 and limited outbreak response and mitigation capabilities may not be sufficient to contain an avian influenza outbreak.
None declared.
None.
This work was funded by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, under contract number HHSN266200700005C. Marinjho Jonduo was supported by a Partnership in Health Research Training Program, funded by Esso Highlands Papua New Guinea Limited.