a Landcare Research, Dunedin, New Zealand.
b Arbovirus Surveillance and Research Laboratory, School of Pathology and Laboratory Medicine, The University of Western Australia, Australia.
c New Zealand Centre for Conservation Medicine, Auckland, New Zealand.
d Investigation and Diagnostic Centres, Wallaceville, Upper Hutt, New Zealand.
e Institute of Agriculture and Environment, Massey University, Palmerston North, New Zealand.
f Institute of Environmental Science and Research, Porirua, New Zealand.
Correspondence to Daniel Tompkins (e-mail:tompkinsd@landcareresearch.co.nz).
To cite this article:
Tompkins D et al. Surveillance for arboviral zoonoses in New Zealand birds.
Western Pacific Surveillance and Response Journal, 2013, 4(4):16–23. doi:10.5365/wpsar.2013.4.3.002
Introduction: Given the significant burden that emerging infectious diseases place on global economies and public health, the monitoring and mitigation of, and early response to, potential infectious diseases are of the highest priority. The objective of this study was to survey for known and other potential arboviral zoonoses in multiple bird species at four locations in New Zealand.
Methods: Common bird species were targeted for blood sampling during two southern hemisphere summers. Sera from each period (n = 185 and n = 693) were screened in an epitope blocking enzyme immunoassay for flavivirus antibody detection. In the first season, testing for antibodies to specific alphaviruses was conducted on samples with sufficient sera (n = 22). In the second season, blood clots (n = 544) were screened for viral presence by polymerase chain reaction (PCR) for alphaviral and flaviviral RNA, and virus isolation (n = 146) was conducted.
Results: Flavivirus antibodies were detected in 13 species, and one Australasian gannet (Morus serrator) from one site was positive for antibodies to Ross River virus. PCR tests and virus isolation were all negative.
Discussion: Evidence for flavivirus exposure in seabirds at Kaikoura Peninsula and Cape Kidnappers suggests that viruses isolated from seabirds and associated ticks in New Zealand in the late 1970s are still present. Evidence for flavivirus exposure in passerines at Kaikoura Peninsula, Cape Kidnappers and Mokoia Island is novel. The Ross River virus finding is also new and supports the hypothesis that migratory seabirds are an import pathway for such agents into New Zealand.
Emerging infectious diseases (EIDs; disease-causing agents that rapidly increase in host range, geographic range or prevalence) are a well-recognized threat to public health globally,1 and the rate of disease emergence has risen since the middle of the 20th century.2 Risk analysis indicates that emergence is driven by multiple factors including socioeconomic circumstances,2,3 climate and land-use changes,4,5 and pathogen pollution (the anthropogenic global movement of pathogens).6 Given the significant burden that EIDs place on global economies and public health,1,7 the monitoring and mitigation of, and early response to, potential infectious disease threats are of the highest priority.4,8 These global concerns are reflected in New Zealand with an increase in active surveillance for otential disease threats being advocated for the benefit of native wildlife, domestic stock and public health.9–15
Four potential viral zoonoses associated with wildlife have previously been documented in New Zealand: three flaviviruses (Johnston Atoll virus,16,17 Saumarez Reef virus and an unnamed Hughes group virus17) and one alphavirus (Whataroa virus18). The flaviviruses are all tick-borne viruses that have remained largely unstudied since their detection in the late 1970s. Johnston Atoll virus is closely related to the Quaranfil group of viruses, which have been isolated from symptomatic humans,16 and it has been hypothesized that humans may also be susceptible to infection with Johnston Atoll virus.16,19 Saumarez Reef virus is believed to have been responsible for febrile illness in meteorological workers on the Saumarez and Frederick reefs in Australia.20 A closely related Hughes group virus, Soldado virus, has been implicated as a cause of human illness overseas.21 The Whataroa virus is a mosquito-borne alphavirus that belongs to the Sindbis virus subgroup that has had a known public health impact in several countries.22 Whataroa virus has been detected only in bird populations and two endemic mosquito species (Culex pervigilans and Culiseta tonnoiri) to date, around Whataroa township on New Zealand’s South Island.18
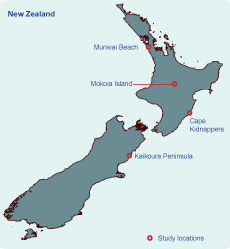
The ecology and host-associations of all four viruses are poorly understood. In this study we conducted wildlife surveillance for these and other potential viral zoonoses at two locations where viruses were previously recorded (Kaikoura Peninsula and Cape Kidnappers; Figure 1) and two locations where occurrence was likely (Muriwai Beach for tick-borne viruses and Mokoia Island for mosquito-borne viruses). These locations are also potential import pathways for infectious agents into New Zealand; for example, migratory seabirds and their ticks may be able to transport infections such as West Nile virus into the country.23 This potential import pathway has been discussed by various researchers globally,24–27 and the risk to New Zealand needs to be determined.

The Kaikoura Peninsula, on the north-east coast of New Zealand’s South Island, is where Saumarez Reef virus and the unidentified Hughes group arbovirus were isolated from ticks associated with both the red-billed gull (Larus novaehollandiae) and white-fronted tern (Sterna striata) colonies in the 1970s and where the Hughes group virus was isolated from the blood of a red-billed gull.17 The presence of these viruses suggests a potential import pathway of migratory seabirds.23 Red-billed gulls can move over 300 km after breeding, with some evidence of trans-oceanic straggling.28 Large numbers of white-fronted terns migrate from New Zealand to Australia; the farthest recovery of a banded bird was 2970 km from Kaikoura to South Australia (Figure 2).28
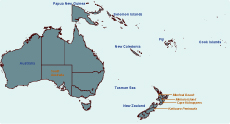
Cape Kidnappers, a peninsula on the east coast of New Zealand’s North Island, has the country’s largest mainland colony of the migratory Australasian gannet (Morus serrator). In the 1970s, Johnston Atoll virus was isolated from ticks associated with these gannets, in addition to the unidentified Hughes group arbovirus also isolated on the Kaikoura Peninsula.16,17 Most young Australasian gannets cross the Tasman Sea within three months of life,28 remain in Australian waters until they are two to three years old (Figure 2), then return to their natal gannetries at three years of age as non-breeding or roosting birds – another potential import pathway.
Muriwai Beach, on the west coast north of Auckland, has three potential import pathways. First, it is a second mainland colony site for migratory Australasian gannets; second, the site is close to major shipping ports and airports in the Auckland Region (both potential sites of entry of exotic vectors); and third, it is a popular tourist destination attracting thousands of overseas visitors each year. Being in the north of the country it also has close proximity to Australia and the Pacific islands (Figure 2).
Mokoia Island is a 1.35 km2 island in the middle of Lake Rotorua in the centre of New Zealand’s North Island. Infection of local bird populations by mosquito-borne avian malarial parasites have been documented here,29 making it a potential site for mosquito-borne viral agents such as Whataroa virus. In addition, the migration of shining cuckoos (Chrysococcyx lucidus; a species that breeds on Mokoia Island) to the Bismarck (New Britain Island) and Solomon archipelagos and other Pacific Islands28,30 offers a potential route of agent incursion (Figure 2). Mokoia Island is used for endangered bird translocations, representing a pathway for viral spread within the country.
The common bird species present at each site were targeted for blood sampling during two southern hemisphere summers – January to March 2008 (all four sites) and November 2008 to February 2009 (Kaikoura Peninsula, Cape Kidnappers and Mokoia Island only). Tuis (Prosthemadera novaeseelandiae), North Island robins (Petroica longipes), North Island saddlebacks (Philesturnus rufusater) and other passerines were caught using mist nets, banded with a numbered metal band (if no band already present) and had a peripheral blood sample collected from the brachial vein. The vein was punctured using a sterile 25–27 g needle (depending on bird size), and blood (no more than 1% body weight) was collected into capillary tubes.
Hand nets were used to catch red-billed gulls and white-fronted terns, and shepherd’s crooks were used to catch Australasian gannets. Little blue penguins (Eudyptula minor) were taken by hand from burrows as were gulls and terns from nests. Wekas (Gallirallus australis) were caught in baited cage-traps, and New Zealand scaup (Aythya novaeseelandiae) were caught in mist nets on the shore of Lake Rotorua (in which Mokoia Island lies). Once banded with a numbered metal band (if no band already present), a peripheral blood sample was collected. Gannets, penguins, gulls, terns, scaups and wekas had up to 1.0 ml blood drawn by syringe with a sterile 25 g needle from the metatarsal vein. Gulls and juvenile terns had their brachial vein punctured using a sterile 25 g needle with up to 0.5 ml blood collected into capillary tubes.
Serum samples (collected from n = 185 and n = 693 individuals during the first and second field seasons respectively) were screened using an flavivirus epitope-blocking enzyme-linked immunosorbent assay described elsewhere31,32 with the exception that virus-inactivated cell culture lysates were used to coat U-bottom 96-well plates before addition of test samples.33 Briefly, after washing excess antigen and blocking, sera were added to the 96-well plates in duplicate before the addition of the flavivirus group-reactive monoclonal antibody 3H6 (JCU Tropical Biotechnology Pty Ltd, Townsville, Australia). Binding of the monoclonal antibody was detected following the addition of horseradish peroxidise-conjugated goat anti-mouse antibody and subsequent visualization of enzymatic activity in substrate buffer. Optical densities were measured and percentage inhibition of the monoclonal antibody by test sera was calculated using negative control sera as the reference. For samples with sufficient sera, those with 30% or greater inhibition were re-tested against 3H6 as well as specific monoclonal antibodies 10C6 (JCU Tropical Biotechnology Pty. Ltd) for Murray Valley encephalitis virus and 3.1112G (Discipline of Microbiology and Immunology, The University of Western Australia, Perth, Australia) for Kunjin virus (both flaviviral agents of incursion concern from Australia14). Samples with 50% or greater inhibition on at least one 3H6 test were considered positive for flavivirus antibodies. This criterion was validated as robust in the 50 samples that were re-tested; while some samples up to 40% did not confirm at re-testing, all samples over 40% did.
Testing for antibodies to specific alphaviruses (Ross River virus, Barmah Forest virus and Sindbis virus; arboviral agents of incursion concern from Australia14,34) was also carried out on first field season samples with sufficient remaining sera (n = 22) using serum neutralization assays as described elsewhere35 except that Vero cells were used in place of baby hamster kidney cells. In short, sera were serially diluted in 96-well tissue culture plates and incubated for five days with approximately 50 tissue culture infectious doses of virus and Vero cells. Each well was examined microscopically for cytopathic effect (CPE), and neutralization titres were expressed as the reciprocal of the highest serum dilution where CPE did not occur. Samples with two repeat neutralization titres of at least 40 were considered positive.
Blood clots collected during the second field season (from n = 544 individuals) were screened for viral presence using flavivirus and alphavirus group-specific reverse transcription–PCR tests. Blood clots collected after removal of serum were frozen at –70 °C within four hours of collection. They were then homogenized in sterile virus transport media and the debris pelleted by microcentrifugation. The collected supernatant was extracted directly using the Zymo Viral RNA kit (Zymo Research Corporation, Irvine, CA, USA) and resuspended in ddH2O. For flavivirus testing, we employed the flavivirus nsp5 PCR that uses mFU1 and cFD2 published primers.36,37 Generic alphavirus PCR was conducted using nsp4 AL-EF and AL-ER primers.38 Both PCR tests were conducted using Invitrogen Superscript III Platinum Taq Sybr Green one-step qRT–PCR master mix (Life Technologies, Carlsbad, CA, USA) in single-tube reactions. The detection of positive reactions was determined by melt curve analysis of the PCR product followed by gel electrophoresis and DNA sequencing of PCR amplicons.
Virus isolation was performed on clots collected from 146 individuals during the second field season. Supernatants from homogenized clots were inoculated into VeroE6 cell cultures39 for two passes of five days and monitored for evidence of CPE using a light microscope.
In the first field season, antibodies to flavivirus were detected in serum samples from a red-billed gull at Kaikoura Peninsula and a North Island saddleback at Mokoia Island (Table 1). In the second field season, a relatively high prevalence of antibodies to flavivirus was observed in serum samples from white-fronted terns at Kaikoura Peninsula (Table 1). Flavivirus antibodies were also detected at this time in red-billed gulls and passerines at this location; in little blue penguins and passerines at Cape Kidnappers; and in passerines, wekas and New Zealand scaups at Mokoia Island (Table 1). None of the 50 repeat-tested samples were specifically positive for either Murray Valley encephalitis virus or Kunjin virus.
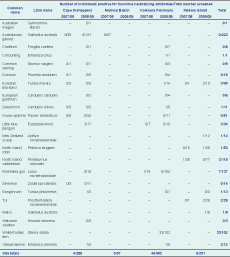
Of the 22 first field season samples also tested for antibodies to specific alphaviruses (Table 2), one Australasian gannet from Muriwai Beach was positive for antibodies to Ross River virus with two repeat neutralization titres of 80.

* Specific alphaviruses – Ross River virus (RRV), Barmah Forest virus, Sindbis virus. See Table 1 for species Latin names.
In the second field season, the 544 blood clots (from Kaikoura Peninsula, Cape Kidnappers and Mokoia Island) screened on alphavirus and flavivirus generic PCR tests were all negative (Table 3). The 146 clots subject to viral isolation were also negative (Table 3); no CPE was observed in any of the cultures after two passages of virus isolation in VeroE6 cells, and no flavivirus PCR products were amplified with RNA extracted from these cell cultures.
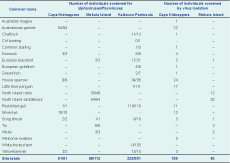
* All tests were negative. See Table 1 for species Latin names.
The four sites surveyed for viral agents in birds were selected on the basis of previous documentation of potential zoonoses (in seabirds and their associated ticks) and/or the presence of potential import pathways. Our results indicate that these selection criteria were relevant. Evidence suggests the continued presence of previously isolated seabird flaviviruses, the presence of novel avian flaviviral agents and exposure of a migratory species to an alphavirus of incursion concern from Australia. This last result, serological evidence for antibodies to Ross River virus (the most common mosquito-borne pathogen causing human disease in Australia34) in an Australasian gannet at Muriwai Beach, is a novel finding of particular relevance to public health.
Although the standard positive criterion for the flavivirus serology conducted is to achieve inhibition of 3H6 on repeat testing, we were frequently unable to obtain sufficient serum for a repeat (particularly from smaller birds). To maximize the utility of our surveys, and prevent biasing against smaller species in our findings, we instead used a criterion of 50% or greater inhibition on at least one test. Although this criterion was validated as robust in the 50 samples that were re-tested (while some samples up to 40% did not confirm at re-testing, all samples over 40% did), our inability to conduct repeat testing on all samples means that cases of just one or two positive results should be interpreted with caution and require follow-up sampling to confirm the evidence for flavivirus infection. In spite of this proviso, we have obtained two strong lines of evidence for such infection.
First, serology results from Kaikoura Peninsula suggest that previously isolated flaviviruses from red-billed gulls (the unidentified Hughes group arbovirus) and ticks associated with both red-billed gulls and white-fronted terns (Saumarez Reef virus and the unidentified Hughes group arbovirus) are still present at this site. Targeted sampling at different times of year may be required for successful viral isolation to verify agent identity. With specific tests for flaviviral agents of incursion concern being negative, the flaviviral reactivity detected in little blue penguins at Cape Kidnappers similarly suggests that the viruses previously isolated from ticks associated with Australasian gannets at this site (Johnston Atoll virus and the unidentified Hughes group arbovirus) may also still be present. However, successful viral isolation is again necessary to verify this.
Second, serological evidence for flavivirus exposure in passerines is novel with no prior evidence for such agents being present in such hosts. Targeted sampling at different times of year may once again be required for successful viral isolation to identify the agents present and inform whether this represents a past incursion via a migratory species such as the shining cuckoo. Since human flaviviral infection is as yet unknown in New Zealand,9 these agents are most likely not a risk to public health.
The key conclusion that can be drawn from both the results discussed above and previous work is that migratory birds represent a possible import pathway for potential zoonotic agents into New Zealand. Both the past and current evidence for Saumarez Reef virus and Johnston Atoll virus support the hypothesis that this pathway has historically operated to bring such agents into the country. Although birds may not be currently carrying viral particles back into New Zealand, the evidence for Australasian gannet exposure to Ross River virus indicates that incursion from Australia by such a mechanism may be possible. Since the native Aedes notoscriptus and Culex pervigilans and the introduced Aedes camptorhynchus, Aedes australis and Culex quinquefasciatus mosquitoes are all potentially competent vectors of Ross River virus,14,40 such incursion could lead to ongoing transmission within the country. With this agent being of public health concern, more thorough surveillance should be carried out at Muriwai Beach to confirm its current status.
None declared.
This work was funded by the New Zealand Foundation for Research Science and Technology (now Ministry of Business, Innovation and Employment).
Thanks to Lindsay Rowe and Jim Mills for field assistance at Kaikoura Peninsula, to Raewyn Edmonds and Te Rūnanga o Kaikoura for iwi approval to work at Kaikoura Peninsula, to Mick Unahi and Ngāti Hawea for iwi approval to work at Cape Kidnappers, to Malcolm Paterson and Ngāti Whatua o Kaipara for iwi approval to work at Muriwai Regional Park, and to Bill Kingi and the Mokoia Island Trust for iwi approval to work on Mokoia Island. Thanks also to Dean Clarke, Morgan Coleman, Keven Drew, Steph Hicks, Pete Lei, Adrian Monks, Maria Barclay, Lauren Best, Kirsten Derry, Mel Farrant, John Potter, Stephanie Shaw, Ellen Schoener, Cleland Wallace, Stefanie Ismar and Katja Geschke for field assistance. Further thanks to Della Orr for help with virology test development, Megan Dymond and Jianning Wang for contributions to PCR test development and Cheryl Johansen for serological testing. This work was conducted under New Zealand Department of Conservation (DOC) Global Concession CA-5160-OTH; DOC Research and Collection Permits NM-22225-RES, ECHB-22299-FAU, AK-22099-FAU, BP-22190-RES, NM-23980-RES, ECHB-24005-FAU and BP-23988-RES; Landcare Research Animal Ethics Authority 07/12/01; New Zealand National Bird Banding Scheme Institutional Permit to Band Birds No. 2007/83.