A new paper by a team of University of Notre Dame researchers that included Shahriar Mobashery, Jeffrey Peng, Brian Baker and their researchers Oleg Borbulevych, Malika Kumararasiri, Brian Wilson, Leticia Llarrull, Mijoon Lee, Dusan Hesek and Qicun Shi describes a unique process that is central to induction of antibiotic resistance in the problematic bacterium methicillin-resistant Staphylococcus aureus (MRSA).
MRSA first emerged in the United Kingdom in 1961 and spread rapidly across the globe. Modern strains of MRSA are broadly resistant to antibiotics of various classes, but resistance to β-lactam antibiotics, which include penicillins, cephalosporins, and carpapenems, is an acute problem because it impacts virtually all commercially available members of the class.
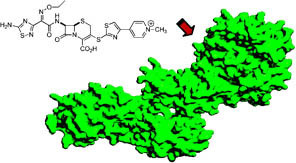
Earlier research by Mobashery, who holds the Navari Family Chair of Life Sciences at Notre Dame, found that an antibiotic sensor/signal transducer protein called “BlaR1” is a key player in MRSA’s resistance to β-lactam antibiotics. Specifically, he had detected by spectroscopy a unique recognition process by the BlaR1 protein of the antibiotic that the organism might encounter. This recognition event, termed “Lysine N-Decarboxylation Switch,” involved formation of an N-carboxylated lysine within the antibiotic-binding domain of BlaR1, which experiences decarboxylation on binding to the antibiotic. This decarboxylation gives the antibiotic complex longevity, which benefits MRSA in the face of the antibiotic challenge. Although this antibiotic-recognition event was described by Mobashery’s research group a few years earlier, the process was not visualized in atomic resolution, despite attempts by several other research groups.

The three collaborating groups of Notre Dame researchers approached the problem differently. The Peng group studied the process by three- and two-dimensional NMR spectroscopy in Notre Dame’s Lizzardo Magnetic Resonance Research Center, the Baker group grew new crystals of the protein for x-ray diffraction and the Mobashery group applied computational methods to understand the process.
The efforts paid off, as Peng demonstrated the presence of N-carboxylated lysine in the protein and showed that it undergoes N-decarboxylation on binding to the antibiotic in solution. Baker visualized both the N-carboxylated lysine in the x-ray crystal structure of the uncomplexed form and showed that when the antibiotic complexed with the protein, the N-decarboxylation switch resulted in a stable complex critical to the manifestation of resistance.
The lysine N-decarboxylation switch triggers MRSA’s antibiotic sensor domain to adopt the active state that leads to all the subsequent biochemical processes that enable resistance, an event that was investigated by computational analyses in the Mobashery lab in the present study.
The importance of this lysine N-decarboxylation switch for MRSA rests in the fact that the organism does not mobilize its resources until and unless it is exposed to the antibiotic. As such, in an economy of existence, the MRSA conserves its resources until BlaR1 informs it that it has come in contact with a β-lactam antibiotic.
The research paper describing the team’s findings appears in the Journal of Biological Chemistry.
Contact: Shahriar Mobashery, 574-631-2932, mobashery@nd.edu to:mobashery@nd.edu