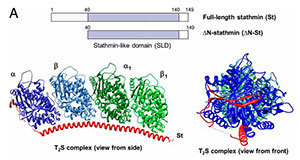
 Schematic showing the human stathmin constructs used in this work (Click for larger view)
Schematic showing the human stathmin constructs used in this work (Click for larger view)
A new research paper from the labs of University of Notre Dame researchers Holly Goodson and Mark Alber helps resolve an ongoing debate about the assembly of a subcellular network that plays a critical role in cell function and disease.
Goodson and her former postdoctoral fellow Kamlesh Gupta (now a senior scientist at W. M. Keck Center for Transgene Research) from the Department of Chemistry and Biochemistry teamed up with Alber’s group from the Department of Applied and Computational Mathematics and Statistics to study the dynamical behavior of subcellular fibers called microtubules. The microtubule cytoskeleton is a dynamic polymer network that plays a crucial role in cell division, assembling into the remarkable machine that partitions the DNA. It also forms a transport network that helps cells distribute nutrients and building materials.
“This fiber network is analogous to a railway system, with the microtubules acting as rails for molecular engines that move cargo containers around the cell,” Goodson said. “However, unlike human railway systems, which are stable over time, the microtubules are constantly being laid down and picked up.”
The constant turnover of these structures is important because it enables the transport network to find its cargo and rearrange in response to cell movements and division. Because of its significance for cell function, this microtubule turnover process is the target of some key anticancer drugs.
The microtubule assembly and dynamics are precisely controlled, and a key regulator is the microtubule destabilizer known as stathmin. Stathmin’s precise method of action has been open to debate and has remained controversial. One proposed model is that it reduces polymer indirectly by sequestering microtubule units. Another model is that stathmin acts directly on microtubules by an as-yet-unknown mechanism.
The new paper by the Goodson and Alber groups provides a resolution to this debate by explaining how stathmin works. The experiments, primarily designed and performed by Gupta, present experimental evidence that stathmin can act directly on microtubules, and it does so by binding and destabilizing segments of the assembling microtubule before they can be incorporated into the final microtubule structure. Accompanying computer simulations show that this type of molecular activity could produce the experimentally observed effects of microtubule dynamics.
“This work is significant because this disassembly process is essential for basic cell survival and because stathmin, also called oncoprotein 18, is dramatically overproduced in a number of cancers,” Alber said. “Understanding how the protein works is an important step toward figuring out how to inhibit it, which may provide a route for new anticancer drugs.”
The research effort combined Goodson’s expertise in biochemistry and cell biology and Alber’s expertise in mathematical and computational biology, resulting in an interdisciplinary approach that used both experiments and computer simulations. The researchers are members of Notre Dame’s Center for the Study of Biocomplexity. The center’s researchers come from different departments in the Colleges of Science and Engineering and are working together to meld physical, mathematical and computational approaches with those of modern biology to understand biomedical problems in a quantitative and predictive way.
The collaboration’s National Science Foundation-funded paper appeared last month in the Proceedings of the National Academy of Sciences (PNAS).