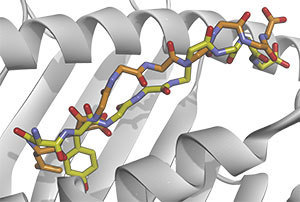
 Notre Dame researchers and their collaborators explain how identifying distinctions between mutant (yellow) and normal (orange) immune targets can help locate neo-epitopes that elicit anti-cancer immune responses
Notre Dame researchers and their collaborators explain how identifying distinctions between mutant (yellow) and normal (orange) immune targets can help locate neo-epitopes that elicit anti-cancer immune responses
A team of University of Notre Dame scientists, in collaboration with researchers at the University of Connecticut, have announced the results of a new study on identifying potential targets for personalized cancer vaccines. The paper, “Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity,” was recently published in the Journal of Experimental Medicine. The research group at Notre Dame was led by Brian Baker, associate dean for research and graduate studies and professor of chemistry and biochemistry, and included Steven Corcelli, associate professor of chemistry and biochemistry, and graduate student Cory Ayers.
“There has been a lot of attention on cancer vaccines in the last 10 years. The potential is huge, but actual progress has been slow,” said Baker.
Vaccines are introduced into the body to create or boost an immune response to protein antigens that differ from those normally produced by the host, such as those from viral proteins. Tumors develop due to mutations in a person’s DNA, causing production of mutated proteins and neo-antigens that also differ from those produced by the host. “Many of these neo-antigens result in the destruction of early-stage cancerous cells by the immune system,” said Baker. “In cases of established cancer, however, the immune response is insufficient, but can be boosted through vaccines.” It has been difficult for researchers to develop vaccines for cancer based on neo-antigens because they are rare and can vary from person to person. Additionally, identifying those neo-antigens that differ enough from the host to elicit an actual immune response presents yet another challenge.
 Brian Baker
Brian Baker
The study discusses two methods of identifying, with relatively high accuracy, potential neo-antigens for use as personalized cancer vaccines. Combing high-throughput genome sequencing and bioinformatics, the University of Connecticut team identified the neo-antigens present in mouse models of cancer. The Notre Dame group then combined the sequencing data with three-dimensional structural modeling to predict which neo-epitopes differed most from those present in the normal mouse genome, helping identify the neo-epitopes that were the most immunogenic.
“The next step is to repeat this methodology with tumors from human cancer patients,” explained Baker. “Our goal is to one day see personalized vaccines play a starring role in cancer therapy. With advances in DNA sequencing and computational modeling, the process could be done quickly — from the time the tumor genome is sequenced and analyzed, to the production of a vaccine cocktail — it could be done in a matter of weeks.”
“This study is hugely exciting because it combines cancer immunology with personalized medicine, which are two of the most exciting areas of biomedical research right now,” Baker said. The research groups have submitted an application for a clinical trial with ovarian cancer patients and expect to begin the trial this winter. Further highlighting the impact of Notre Dame science, the trial will be led by Notre Dame alumnus Dr. Angie Kueck of the University of Connecticut Carole and Ray Neag Comprehensive Cancer Center. It will be among the first in the world to test genomics-driven, personalized cancer immunotherapy, and the first of its kind for ovarian cancer.
Contact: Brian Baker, 574-631-9810, brian-baker@nd.edu