A new tool may allow researchers to see more of the physiological state of living organisms at the cellular level, according to a study by the University of Notre Dame.
Published in Development, the study shows how an open-source application, created by Notre Dame researchers, can use two different conventional microscope images obtained at low excitation powers to create one high-resolution, three-dimensional image. The application, dubbed DeSOS, combines imaging techniques used within the program: blind deconvolution (De), which allows for the recovery of blurred images in certain circumstances, and stepwise optical saturation super-resolution (SOS), an imaging method that helps extend the resolution beyond its typical diffraction limit.
In full, DeSOS uses physics to identify differences between the two uploaded images and produce one image with significantly greater clarity than previously possible with standard lab equipment.
“This open-source application permits scientists to extract an image beyond the resolution that they could previously achieve by using equipment they likely already have, a confocal or two-photon microscope,” said Scott Howard, associate professor of electrical engineering and co-lead author on the study. “Not only does this eliminate the expense of purchasing a super-resolution microscope, but our program is also faster and has more functionality than a super-resolution microscope when evaluating living organisms.”
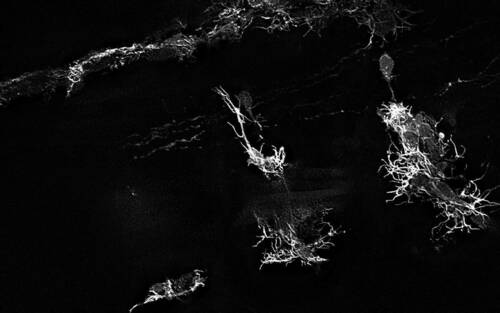
For the study, co-lead author Cody J. Smith, Elizabeth and Michael Gallagher Assistant Professor of Biological Sciences, used DeSOS to improve imaging for his research on the development of spinal cords. Smith and his team analyzed specialized cells that grow and extend to form the nervous system of zebrafish. The images developed through the application allowed the researchers to see the structures of the zebrafish’s nervous system considerably better than traditional images taken on confocal and two-photon microscopes.
“Because of DeSOS, my lab was able to see individual neuronal fibers during the zebrafish spinal cord development process, including components of our research we couldn’t clearly recognize without this technology,” said Smith. “My lab was able to significantly boost our imaging capabilities and make new discoveries by incorporating this open-source application in our research.”
Beyond still images, this DeSOS technique allows users to compile videos of time-lapsed images from living organisms. In the study, Howard and Smith created a video that showed a zebrafish’s neuronal fibers growing to form branches during development, eventually extending toward the head and posterior to form the nervous system.
The first authors on the paper are Evan Nichols, undergraduate student of neuroscience, and Yide Zhang, graduate student of electrical engineering and Advanced Diagnostics and Therapeutics Berry Family Foundation Graduate Fellow. Other collaborators include Cody Kankel, high performance computing administrator at the Center for Research Computing; Abigail Zellmer, undergraduate student of biological sciences; and Siyuan Zhang, Dee Associate Professor of Biological Sciences.
This study was supported by Notre Dame’s Freimann Life Sciences Center and the Center for Stem Cells and Regenerative Medicine. Funding was provided by the Alfred P. Sloan Foundation, Elizabeth and Michael Gallagher Family, National Institutes of Health the and the National Science Foundation.
Contact: Brandi Klingerman, research communications specialist, Notre Dame Research, research.nd.edu, @UNDResearch
Originally published by at research.nd.edu on Feb. 28.